# Import necessary module
import geopandas as gpd
# Set filepath relative to your `geopython2023` working directory, from
# where your Jupyter Notebook also should be started
fp = "../files/data/L2/DAMSELFISH_distributions.shp"
# depending if you have your notebook file (.ipynb) also under L2
# fp = "DAMSELFISH_distributions.shp"
# or full path for Windows with "r" and "\" backslashes
# fp = r"C:\Users\kmoch\geopython2023\L2\DAMSELFISH_distributions.shp"
# Read file using gpd.read_file()
data = gpd.read_file(fp)Introduction to Geopandas
Downloading data
For this lesson we are using data that you need to download from the provided link above. Once you have downloaded the damselfish-data.zip file into your geopython2023 directory (ideally under L2), you can unzip the file using e.g. 7Zip (on Windows).
DAMSELFISH_distributions.dbf DAMSELFISH_distributions.prj DAMSELFISH_distributions.sbn DAMSELFISH_distributions.sbx DAMSELFISH_distributions.shp DAMSELFISH_distributions.shp.xml DAMSELFISH_distributions.shx
The data includes a Shapefile called DAMSELFISH_distribution.shp (and files related to it).
Reading a Shapefile
Spatial data can be read easily with geopandas using gpd.read_file() -function:
Let’s see what datatype is our ‘data’ variable
print(type(data))<class 'geopandas.geodataframe.GeoDataFrame'>So from the above we can see that our data -variable is a GeoDataFrame. GeoDataFrame extends the functionalities of pandas.DataFrame in a way that it is possible to use and handle spatial data within pandas (hence the name geopandas). GeoDataFrame have some special features and functions that are useful in GIS.
Let’s take a look at our data and print the first 5 rows using the head() -function prints the first 5 rows by default
display(data.head(5))| ID_NO | BINOMIAL | ORIGIN | COMPILER | YEAR | CITATION | SOURCE | DIST_COMM | ISLAND | SUBSPECIES | ... | RL_UPDATE | KINGDOM_NA | PHYLUM_NAM | CLASS_NAME | ORDER_NAME | FAMILY_NAM | GENUS_NAME | SPECIES_NA | CATEGORY | geometry | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 183963.0 | Stegastes leucorus | 1 | IUCN | 2010 | International Union for Conservation of Nature... | None | None | None | None | ... | 2012.1 | ANIMALIA | CHORDATA | ACTINOPTERYGII | PERCIFORMES | POMACENTRIDAE | Stegastes | leucorus | VU | POLYGON ((-115.64375 29.71392, -115.61585 29.6... |
| 1 | 183963.0 | Stegastes leucorus | 1 | IUCN | 2010 | International Union for Conservation of Nature... | None | None | None | None | ... | 2012.1 | ANIMALIA | CHORDATA | ACTINOPTERYGII | PERCIFORMES | POMACENTRIDAE | Stegastes | leucorus | VU | POLYGON ((-105.58995 21.89340, -105.56483 21.8... |
| 2 | 183963.0 | Stegastes leucorus | 1 | IUCN | 2010 | International Union for Conservation of Nature... | None | None | None | None | ... | 2012.1 | ANIMALIA | CHORDATA | ACTINOPTERYGII | PERCIFORMES | POMACENTRIDAE | Stegastes | leucorus | VU | POLYGON ((-111.15962 19.01536, -111.15948 18.9... |
| 3 | 183793.0 | Chromis intercrusma | 1 | IUCN | 2010 | International Union for Conservation of Nature... | None | None | None | None | ... | 2012.1 | ANIMALIA | CHORDATA | ACTINOPTERYGII | PERCIFORMES | POMACENTRIDAE | Chromis | intercrusma | LC | POLYGON ((-80.86500 -0.77894, -80.75930 -0.833... |
| 4 | 183793.0 | Chromis intercrusma | 1 | IUCN | 2010 | International Union for Conservation of Nature... | None | None | None | None | ... | 2012.1 | ANIMALIA | CHORDATA | ACTINOPTERYGII | PERCIFORMES | POMACENTRIDAE | Chromis | intercrusma | LC | POLYGON ((-67.33922 -55.67610, -67.33755 -55.6... |
5 rows × 24 columns
Let’s also take a look how our data looks like on a map. If you just want to explore your data on a map, you can use .plot() -function in geopandas that creates a simple map out of the data (uses matplotlib as a backend):
# import matplotlib, make it show plots directly in Jupyter notebooks
import matplotlib.pyplot as plt
data.plot()<Axes: >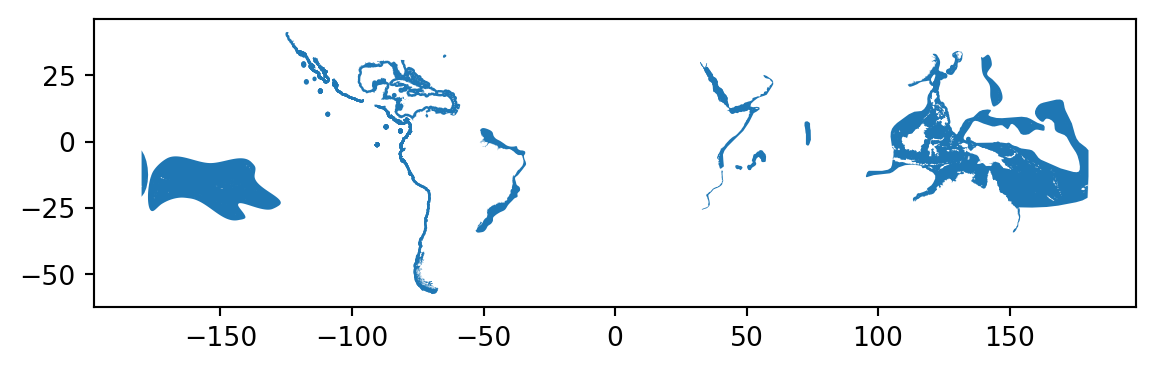
Writing a spatial datafile
Writing a layer into a spatial data file is something that is needed frequently.
Typical spatial vector file formats are:
- GeoPackage (file extension: .gpkg, driver=“GPKG”, layer=“layername”)
- Shapefile (file extension: .shp + several more, default)
- GeoJSON (file extension: .geojson, driver=“GeoJSON”, only use for web maps)
Let’s select 50 first rows of the input data and write those into a new Shapefile by first selecting the data using index slicing and then write the selection into a Shapefile with the .to_file() -function on a GeoDataFrame:
# Create a output path for the data
out_file_path = "DAMSELFISH_distributions_SELECTION.shp"
# Select first 50 rows, this a the numpy/pandas syntax to ``slice`` parts out a dataframe or array, from position 0 until (excluding) 50
selection = data[0:50]
# Write those rows into a new Shapefile (the default output file format is Shapefile)
selection.to_file(out_file_path)Task: Open the Shapefile now in QGIS (or ArcGIS) on your computer, and see how the data looks like.
Geometries in Geopandas
Geopandas takes advantage of Shapely’s geometric objects. Geometries are typically stored in a column called geometry (or geom). This is a default column name for storing geometric information in geopandas.
Let’s print the first 5 rows of the column ‘geometry’:
# It is possible to use only specific columns by specifying the column
# name within square brackets []
data['geometry'].head()0 POLYGON ((-115.64375 29.71392, -115.61585 29.6...
1 POLYGON ((-105.58995 21.89340, -105.56483 21.8...
2 POLYGON ((-111.15962 19.01536, -111.15948 18.9...
3 POLYGON ((-80.86500 -0.77894, -80.75930 -0.833...
4 POLYGON ((-67.33922 -55.67610, -67.33755 -55.6...
Name: geometry, dtype: geometrySince spatial data is stored as Shapely objects, it is possible to use all of the functionalities of Shapely module that we practiced earlier.
Let’s print the areas of the first 5 polygons:
# Make a selection that contains only the first five rows
selection = data[0:5]We can iterate over the selected rows using a specific .iterrows() -function in (geo)pandas and print the area for each polygon:
As stated in the last session, area (or length) calculations purely based on WGS84 latitude/longitude (aka GPS) is not useful as such. To calculate areas reliably we would have to reproject the data into projected coordinate reference system. This will be introduced in the next lessons.
for index, row in selection.iterrows():
# Calculate the area of the polygon
poly_area = row['geometry'].area
# Print information for the user
print("Polygon area at index {0} is: {1:.3f}".format(index, poly_area))Polygon area at index 0 is: 19.396
Polygon area at index 1 is: 6.146
Polygon area at index 2 is: 2.697
Polygon area at index 3 is: 87.461
Polygon area at index 4 is: 0.001Hence, as you might guess from here, all the functionalities of Pandas are available directly in Geopandas without the need to call pandas separately because Geopandas is an extension for Pandas.
Let’s next create a new column into our GeoDataFrame where we calculate and store the areas individual polygons. Calculating the areas of polygons is really easy in geopandas by using GeoDataFrame.area attribute:
data['area'] = data.areaHere your notebook will likely show a warning message:
UserWarning: Geometry is in a geographic CRS. Results from 'area' are likely incorrect. Use 'GeoSeries.to_crs()' to re-project geometries to a projected CRS before this operation.
This is helpful and just reiterates the point, how important it is to consider in which coordinate reference system (CRS) your data is.
Let’s see the first 2 rows of our ‘area’ column.
display(data['area'].head(2))0 19.396254
1 6.145902
Name: area, dtype: float64So we can see that the area of our first polygon seems to be 19.39 and 6.14 for the second polygon. They correspond to the ones we saw in previous step when iterating rows, hence, everything seems to work as it should. Let’s check what is the min and the max of those areas using familiar functions from our previous Pandas lessions.
# Maximum area
max_area = data['area'].max()
# Mean area
mean_area = data['area'].mean()
print("Max area: {:.2f}\nMean area: {:.2f}".format(round(max_area, 2), round(mean_area, 2)))Max area: 1493.20
Mean area: 19.96So the largest Polygon in our dataset seems to be 1494 square decimal degrees (~ 165 000 km2) and the average size is ~20 square decimal degrees (~2200 km2).
Creating geometries into a GeoDataFrame
Since geopandas takes advantage of Shapely geometric objects it is possible to create a Shapefile from a scratch by passing Shapely’s geometric objects into the GeoDataFrame. This is useful as it makes it easy to convert e.g. a text file that contains coordinates into a Shapefile or other geodata format, such as GeoPackage.
Let’s create an empty GeoDataFrame.
# Import necessary modules first
import pandas as pd
import geopandas as gpd
from shapely.geometry import Point, Polygon
# Create an empty geopandas GeoDataFrame
newdata = gpd.GeoDataFrame()
# Let's see what's inside
display(newdata)The GeoDataFrame is empty since we haven’t placed any data inside.
Let’s create a new column called geometry that will contain our Shapely objects:
# Create a new column called 'geometry' to the GeoDataFrame
newdata['geometry'] = None
# Let's see what's inside
display(newdata)| geometry |
|---|
Here will likely also show a warning about an empty Geometry column:
FutureWarning: You are adding a column named 'geometry' to a GeoDataFrame constructed without an active geometry column. Currently, this automatically sets the active geometry column to 'geometry' but in the future that will no longer happen. Instead, either provide geometry to the GeoDataFrame constructor (GeoDataFrame(... geometry=GeoSeries()) or use 'set_geometry('geometry')' to explicitly set the active geometry column.
Typically, we will assign the geometry column through a more elaborate compute function, but here we will do it manually.
Now we have a geometry column in our GeoDataFrame but we don’t have any data yet.
Let’s create a Shapely Polygon representing the Tartu Townhall square that we can insert to our GeoDataFrame:
# Coordinates of the Tartu Townhall square in Decimal Degrees
coordinates = [(26.722117, 58.380184), (26.724853, 58.380676),
(26.724961, 58.380518), (26.722372, 58.379933)]
# Create a Shapely polygon from the coordinate-tuple list
poly = Polygon(coordinates)
# Let's see what we have
display(poly)So now we have appropriate Polygon -object.
Let’s insert the polygon into our ‘geometry’ column in our GeoDataFrame:
# Insert the polygon into 'geometry' -column at index 0
newdata.loc[0, 'geometry'] = poly
# Let's see what we have now
display(newdata)| geometry | |
|---|---|
| 0 | POLYGON ((26.72212 58.38018, 26.72485 58.38068... |
Now we have a GeoDataFrame with Polygon that we can export to a Shapefile.
Let’s add another column to our GeoDataFrame called Location with the text Tartu Townhall Square.
# Add a new column and insert data
newdata.loc[0, 'Location'] = 'Tartu Townhall Square'
# Let's check the data
display(newdata)/tmp/ipykernel_16208/3412220010.py:2: FutureWarning: Setting an item of incompatible dtype is deprecated and will raise in a future error of pandas. Value 'Tartu Townhall Square' has dtype incompatible with float64, please explicitly cast to a compatible dtype first.
newdata.loc[0, 'Location'] = 'Tartu Townhall Square'| geometry | Location | |
|---|---|---|
| 0 | POLYGON ((26.72212 58.38018, 26.72485 58.38068... | Tartu Townhall Square |
Now we have additional information that is useful to be able to recognize what the feature represents.
Before exporting the data it is useful to determine the coordinate reference system (projection) for the GeoDataFrame.
GeoDataFrame has a property called .crs that (more about projection on next tutorial) shows the coordinate system of the data which is empty (None) in our case since we are creating the data from the scratch:
print(newdata.crs)NoneLet’s declare the crs for our GeoDataFrame.
# Set the GeoDataFrame's coordinate system to WGS84
newdata.crs = 4326
# Let's see how the crs definition looks like newdata.crsSetting the coordinate reference system (crs) for the GeoDataFrame like this is akin to “Assigning projection” in ArcGIS or QGIS. We are not changing the coordinate values itself, but only the spatial metadata that is associated with the data.
newdata.plot()<Axes: >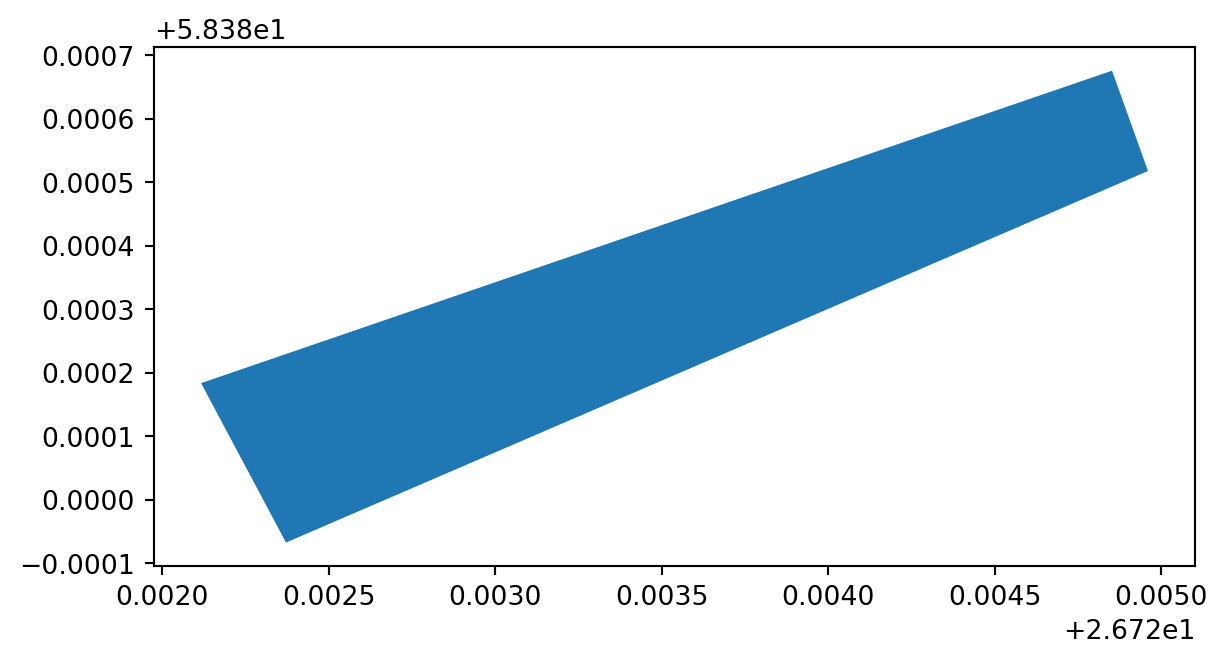
Finally, we can export the data using GeoDataFrames .to_file() -function. The function works similarly as numpy or pandas, but here we only need to provide the output path for the Shapefile:
# Determine the output path for the Shapefile
out_file = "raekoja_plats.shp"
# Write the data into that Shapefile
newdata.to_file(out_file)Now we have successfully created a Shapefile from the scratch using only Python programming. Similar approach can be used to for example to read coordinates from a text file (e.g. points) and create Shapefiles from those automatically.
Task: check the output Shapefile in QGIS and make sure that the attribute table seems correct.
Interactive plotting and data exploration with hvPlot and GeoViews
Since recent versions, GeoPandas support the .explore() function that allows interactive plotting of the data. This function is based on the hvPlot
# lets use the data from the previous example
data.explore()GeoViews is a Python library that makes it easy to explore and visualize geographical datasets.It provides a set of coordinate-aware data types (geometries) and functions for visual integration with the HoloViews library.
GeoViews, like matplotlib, has a large number of options to customize the plots. These will be shown in later lessons. For now, this only serves an example for more interactive plotting in the Jupiter notebook for your convenience, while you explore the datasets.
import geoviews as gv
import geoviews.feature as gf
from cartopy import crs
from bokeh.plotting import figure, output_file, show
gv.extension('bokeh')
data_view = gv.Polygons(data, vdims=['ID_NO', 'BINOMIAL']).opts(tools=['hover'])# this will plot the data in the notebook
(gf.coastline * data_view).opts(width=700, height=400, projection=crs.EckertIV(), global_extent=True)