Lesson 1: Introduction to R
This is the introductory lesson of the course focusing on core R concepts, setting up the R environment, grasping data types and structures, managing R packages, handling data import and export, and performing essential data wrangling and manipulation tasks for spatial data analysis. These foundational skills prepare students for the subsequent advanced lessons, which focus on applying R to geospatial data processing, analysis, and visualization.
Learning outcomes
At the end of this lesson, the students should be able to:
- Configure the R environment and interact with the RStudio interface.
- Gain insight into different data types and structures in R.
- Join attribute tables and conduct essential data wrangling and manipulation tasks.
- Install and load different packages from different sources.
- Import and export various datasets using different packages.
1.1. Setting up the R environment
Setting up the R environment involves the installation, configuration, and preparation of the R along with RStudio on your computer. RStudio is an integrated development environment (IDE) for R that offers tools and features to facilitate R code development, data analysis, and visualization. Downloading and installing R and RStudio is a simple and straightforward process.
To install R:
- Go to this link
- Click on
Download Rto access the download page. - Click on
base - Click on the Download link (this default link will lead you to the latest version, if you desire earlier versions, click on
Previous releases) - Once the download is complete, proceed to install R just as you would install any other software.
To install RStudio:
- Go to this link
- Scroll down to
Installers for Supported Platformsnear the bottom of the page. - Click on the download link corresponding to your computer’s operating system.
- Once the download is complete, proceed to install RStudio just as you would install any other software.
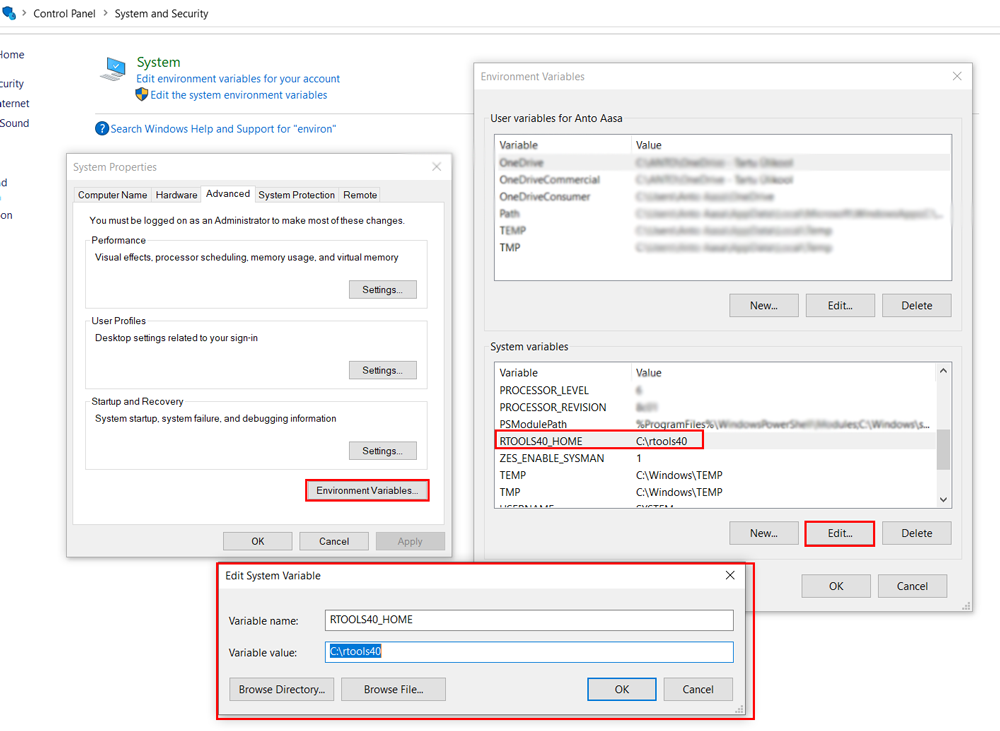
If you are using the Windows operating system, it’s advisable to install RTools. RTools is a collection of tools and utilities for Windows that provides a development environment for building and installing R packages on the Windows operating system. RTools simplifies the process of creating packages and ensures compatibility with the R environment on Windows.
To install RTools on Windows, follow these steps:
- Go to this link
- Click on ‘RTools 4.3 (for
R-4.3.*versions)’ to download the Rtools43 installer. - Once the download is complete, proceed to install RTools just as you would install any other software.
In some cases, updating Rtools in Environmental variables might be needed:
1.2. Getting started with R
Throughout this course, we will be working with R through RStudio. To launch the RStudio, simply click on the RStudio icon or type RStudio at the Windows Run prompt to open it like any other program. After you open RStudio, you should see something like this:
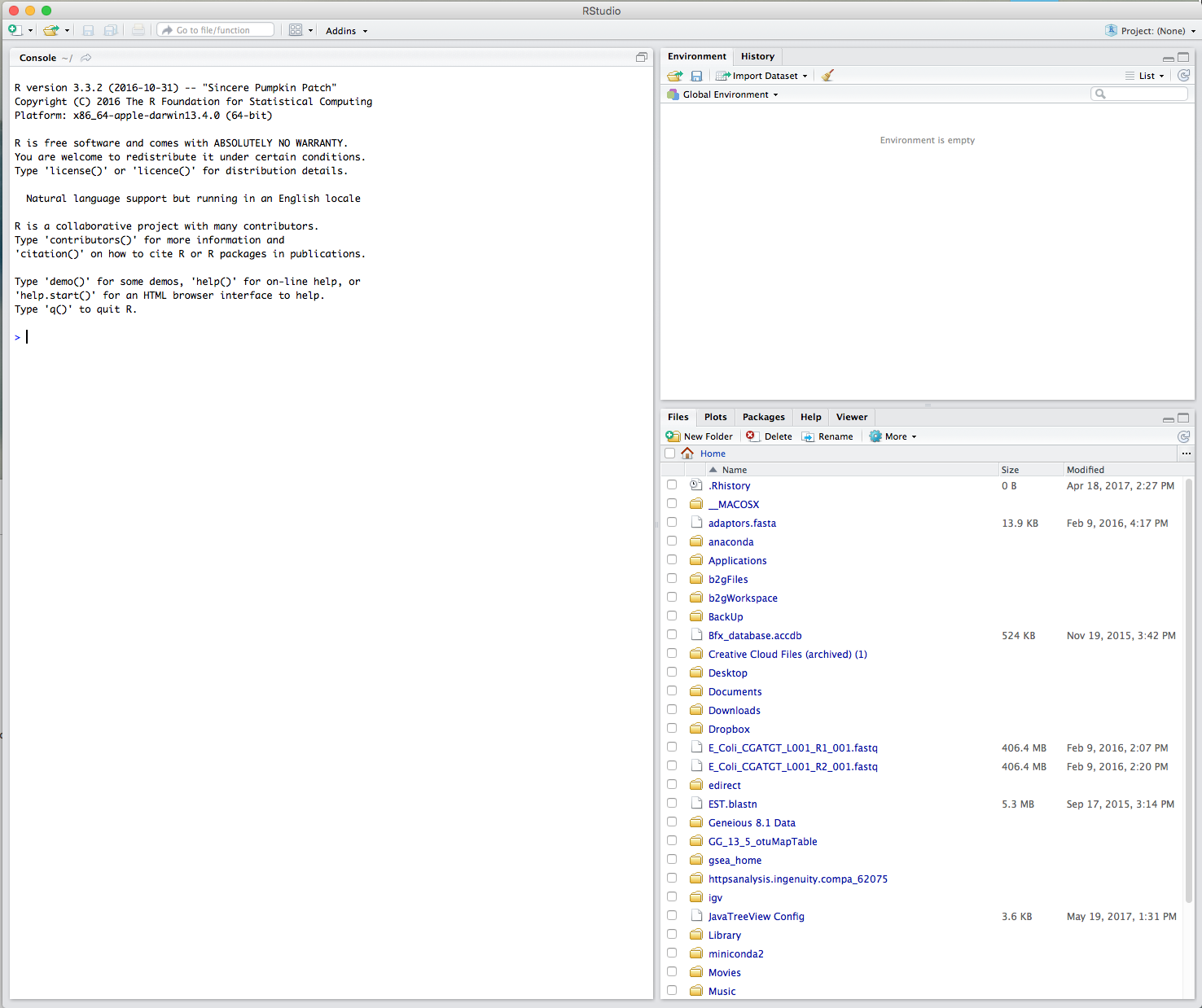
In the default RStudio interface, you could see only three panes dividing the screen: the Console, Files, and Environment panes. To open the Script pane, go to the File menu, then select New File and choose R Script, or use the keyboard shortcut Ctrl + Shift + N. It will create a fourth new pane area in the top-left of the application window, and the Console pane will get shifted down to the bottom-left area.
Let’s start our coding journey by typing some simple statements shown below. To run your code, all you have to do is select the code and then either click on the Run button located on the top-right of the script pane or use the shortcut Ctrl+Enter.
# This is my first code
# Printing a welcome message to the console
print("Welcome to R! R is fun!")[1] "Welcome to R! R is fun!"# Checking the complete version of the installed R
R.version.string # Print the R version string[1] "R version 4.3.1 (2023-06-16)"# Using R as a calculator
sum = 3 + 5 # Perform addition
# Print the result
print(paste("The sum of the numbers is:", sum)) # Display the sum[1] "The sum of the numbers is: 8"# Create a basic scatter plot
plot(1:10)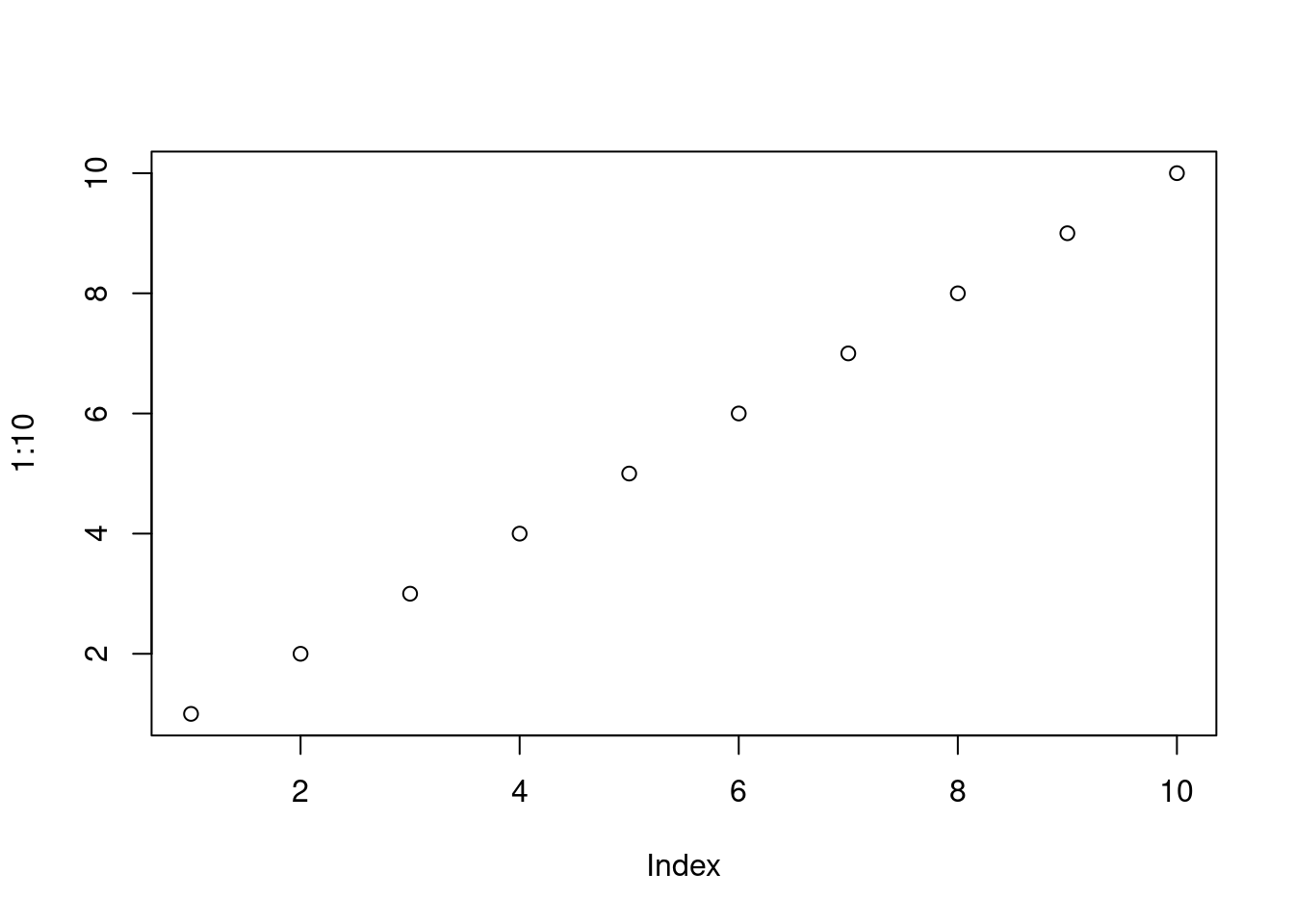
# in the above code chunk is a comment character in R. Anything to the right of a # in a script will be ignored by R. You can easily comment or uncomment an entire chunk of text or code by selecting the desired lines and simultaneously pressing Ctrl + Shift + C on your keyboard.
To stay organized during the lab sessions, we recommend to use your already created course project folder, usually located at C:\Users\yourname\geopython2023, and create new R lesson folders under there, e.g R_01 for lesson 1.
To create the sub-folders in R, we can use dir.create() function. The following code creates a sub-folder named R_01 within geopython2023.
# Create a sub-folder with in geoptyon2023 folder
dir.create("C:/Users/yourname/geopython2023/R_01")To save our code (e.g, the one we created above under This is my first code header), go to the File menu, select Save As..., navigate to the geopython2023\R_01 folder, and save it with a suitable file name.
Once your scripts are saved, you can use the getwd() function to examine the directory where your code has been stored. To observe the files within that directory, you can use the dir() function.
# Check the current working directory
getwd()
# Check the files in the directory
dir() In case you need to read or write files from a specific location outside the geopython2023/R_01 folder, you must set the working directory using the setwd() function and specifying the desired path as an argument as follows:
setwd("Your/Folder/Path")We can also set a working directory manually by clicking on Session menu, then selecting Set as Working Directory, then selecting Choose Directory and navigating to your desired location.
1.3. Creating objects
Creating objects refers to the process of defining and assigning values to variables or objects. R stores everything as an object, including data, functions, models, and output. Creating an object can be done using the assignment operator (<- or =). To activate the assignment operator <-, we can use the keyboard shortcut Alt + - (Windows) and Option + - (Mac).
# Assigning a value to an object
object_name <- value
# Alternatively, you can use '=' for assignment
object_name = valueAlthough both the<-and = operators can be used to assign values to objects, it is recommended to use <- since the = operator can also serve the purpose of passing arguments to functions.
The name of an object reflects the data it holds, making it possible to treat the name as a representation of the information within the object itself.
Using meaningful names can help avoid potential confusion. There are a few rules to keep in mind while naming an object in R:
- Object names should be
short and explicit. - Object names can be created using letters (A-Z, a-z), numbers (0-9), dots (.), or underscores (_), but
cannot start with a number. Datais different thandataas R is case sensitive.- Names of default functions should not be used as object names (
function,if,else,while, etc)
1.4. Data types and data structures
Before diving into specific data processing and analysis tasks, it’s imperative to take some time to examine the data types and data structures in R.
1.4.1. Data types
In R, data types define the type of data that a variable can hold or store. Data types help R understand how to store, manipulate, and process different kinds of information. The class() and typeof() functions are used to determine the data type or class of an object
The most common data types in R include:
Numeric: Numeric data type represents all real numbers with or without decimal values. It is important to note that by default, numeric values are stored in R as double (dbl) unless otherwise specified.
# Create a numeric data object
my_num <- 3.14
my_num[1] 3.14# Check the class name of variable
class(my_num)[1] "numeric"# Check the type of variable
typeof(my_num)[1] "double"# Check if it's numeric
is.numeric(my_num)[1] TRUEIntegers: Integer data type specifies real values without decimal points. We use the suffix L to specify integer data.
# Create an integer object
my_int <- 2
# Check the class
class(my_int)[1] "numeric"Complex: Complex data types represent complex numbers which have both a real and imaginary part. We use the suffix i to specify the imaginary part.
# Create a complex number
my_complex <- 3 + 2i # 2i represents imaginary part
# Check the class
class(my_complex)[1] "complex"Character: Character data type is used to specify character or string values in a variable. Strings are enclosed within single (' ') or double (" ") quotes.
# Create a character data object
my_char <- "Hello, World!"
# Check the class
class(my_char)[1] "character"# Enclosing numbers within quotes also returns character
my_char2 <- "3.5"
# Check the class
class(my_char2)[1] "character"Logical (Boolean): Logical data types represent binary values, which can be either TRUE or FALSE. These are often used for making logical comparisons and decisions.
# Create a logical data object
my_logic <- T
# Check the class
class(my_logic)[1] "logical"Date: Date objects are designed for storing and working with date values. In the code below, we first define the object my_dt_char as a character then use the function as.Date() to convert it to a date. The second argument (%B %d, %Y) defines the date format as entered.
# Define a date string
my_dt_char <- "2023-10-31"
# Convert the date string into a Date object
my_dt_date <- as.Date(my_dt_char)
class(my_dt_date)[1] "Date"# Defines the date format
my_dt_formated <- format(my_dt_date, "%B %d, %Y") # Format the date as "Month Day, Year"
my_dt_formated[1] "October 31, 2023"Raw: Raw data type is used to store binary data as a sequence of bytes. It is a fundamental data type that represents unprocessed, binary data without any specific interpretation. Raw data is often used for tasks such as reading and writing binary files, working with low-level data formats, or encoding data that doesn’t fit neatly into other R data types.
We can use charToRaw() function to convert character data to raw data and rawToChar() function to convert raw data to character data.
# Create a raw vector representing ASCII values for "I love R!"
raw_R <- as.raw(c(73, 32, 76, 111, 118, 101, 32, 82, 33))
raw_R[1] 49 20 4c 6f 76 65 20 52 21# Convert the raw data to a character string
string_R <- rawToChar(raw_R)
string_R[1] "I Love R!"We can convert one type of data to another type of data using as.*() function, where the * represents various data type classes.
as.character(): converts an object to a character data type.as.numeric(): converts an object to a numeric data type.as.integer(): converts an object to an integer data type.as.logical(): converts an object to a logical data type.
Let’s convert some of data types we created earlier to other types:
# Convert numeric to character
num_char <- as.character(my_num)
class(num_char)[1] "character"# Convert character to numeric
char_num <- as.numeric(my_char2)
class(char_num)[1] "numeric"1.4.2. Data structures
Data structure is a way of organizing and storing data in a logical way. In other words, data structure is the collection of data types arranged in a specific order. Selecting the right data structure ensures that data can be effectively utilized, enabling faster and more accurate analysis and computations.
Here are some common types of data structures in R:
Vectors: vectors are the basic data structures in R, representing a one-dimensional array of elements of the same data type. Vectors can be created using the c() function or by coercion from other data types (as.vector()). The is.vector() function checks if an object is a vector.
# Numeric vector
my_vector1 <- c(1, 3, 5, 7, 9)
# Check if "my_vector1" is a vector (should return TRUE)
is.vector(my_vector1)[1] TRUE# Character vector
my_vector2 <- c("A", "B", "c", "D", "E", "F")
class(my_vector1)[1] "numeric"Using square brackets with one or multiple indices enables us to extract (subset) specific values from vectors.
# subsets elements at 2nd and 4th positions
subset_vector <- my_vector1[c(2, 4)]
subset_vector[1] 3 7Lists: lists are is a versatile data structure that can store elements of different data types. They can be nested and contain vectors, data frames, and other lists. Lists are sometimes called recursive vectors, because a list can contain other lists. This makes them fundamentally different from atomic vectors
In R, lists are created using the list() function.
# Create a list with different elements
my_list <- list(c("Red", "Green"), c(21,32,11), TRUE, 51.23, 119.1)
my_list[[1]]
[1] "Red" "Green"
[[2]]
[1] 21 32 11
[[3]]
[1] TRUE
[[4]]
[1] 51.23
[[5]]
[1] 119.1# Convert the lists to vectors using "unlist()" function
list_vec <- unlist(my_list)
list_vec[1] "Red" "Green" "21" "32" "11" "TRUE" "51.23" "119.1"Matrices: Matrices are two-dimensional data structures consisting of rows and columns, where all elements are of the same data type, typically numeric. You can create matrices using the matrix() function by specifying the data elements and the dimensions (rows and columns).
my_matrix <- matrix(c(1, 2, 3, 4, 5, 6, 7, 8, 9), # data elements
nrow = 3, ncol = 3, # dimensions (no. of rows and columns)
byrow = TRUE) # by default matrices are in column-wise order.
my_matrix [,1] [,2] [,3]
[1,] 1 2 3
[2,] 4 5 6
[3,] 7 8 9Data Frames: A data frame is the most common way of storing and displaying tabular data in R. A data frame is two-dimensional data structure similar to a matrix, but store elements of different data types in each column. Data frames are created using the data.frame() function.
# Create a data frame named 'my_df' with columns for id, landuse, description, and year_built
my_df <- data.frame(
ID = c(101, 102, 103, 104),
Landuse = c('Residential', 'Commercial', 'Industrial', 'Park'),
Description = c(
'Residential zone',
'Commercial zone',
'Industrial zone',
'Public park zone'),
Year = c(1990, 2005, 1985, 1970)
)
# Display the 'my_df'
my_df ID Landuse Description Year
1 101 Residential Residential zone 1990
2 102 Commercial Commercial zone 2005
3 103 Industrial Industrial zone 1985
4 104 Park Public park zone 1970# Check the class of 'my_df'
class(my_df)[1] "data.frame"The cbind() and rbind() functions are used to combine data frames. When combining column-wise, the number of rows must match, but row names are ignored. When combining row-wise, both the number and names of columns must match. Use plyr::rbind.fill() to combine data frames that don’t have the same columns.
Let’s create a new data frame called my_df2 with one row and four columns, and then append it to my_df:
# Create a data frame called my_df2
my_df2 <- data.frame(
Area = c(450, 200, 150, 75)
)
# Combine my_df and my_df2 by columns
my_df3 <- cbind(my_df, my_df2)
# Combine df and df2 by rows
my_df4 <- plyr::rbind.fill(my_df, my_df2) # ?plyr::rbind.fillData frame subsetting or the process of extracting elements from a data frame is a frequent task in spatial data analysis. We can access particular rows (observations) or columns in a data frame by employing square brackets [] and the dollar $ operator.
Let’s extract some elements from our combined data frame (my_df3):
# Access a column containing "Landuse" using the $ operator
my_df3$Landuse[1] "Residential" "Commercial" "Industrial" "Park" # Access the cell in the third row and fourth column
my_df3[3, 4] [1] 1985# Filter rows where Area is equal to 200
my_df3[my_df3$Area == 200,] ID Landuse Description Year Area
2 102 Commercial Commercial zone 2005 200# Filter rows where Area is between 100 and 200
my_df3[my_df3$Area > 100 & my_df3$Area < 200,] ID Landuse Description Year Area
3 103 Industrial Industrial zone 1985 150# Filter rows where Area is either greater than 100 or less than 200
my_df3[my_df3$Area > 100 | my_df3$Area < 200,] ID Landuse Description Year Area
1 101 Residential Residential zone 1990 450
2 102 Commercial Commercial zone 2005 200
3 103 Industrial Industrial zone 1985 150
4 104 Park Public park zone 1970 75# Note: a conditional statement based on the & (logical and) operator requires that both conditions are met, however, the | (logical or) operator requires that at least one of the conditions is met.The $ symbol in R is used to access and extract components (such as variables or columns) from data structures, particularly data frames and lists.
Factors: Factors are a specific type of vectors designed for categorical variables with levels. We can create a factors by using the factor() function.
Let’s extract the ‘Landuse’ column from my_df3 and transform it into a factor:
# Convert 'Landuse' column to a factor
my_factor <- factor(my_df3$Landuse)
# Display the attributes
attributes(my_factor)$levels
[1] "Commercial" "Industrial" "Park" "Residential"
$class
[1] "factor"Similar to our approach with data types, we can also convert one type of data structure to another using as.*() function, where in this case * represents data structure classes.
as.data.frame(): converts an object to a data frame.as.list(): converts an object to a list.as.matrix(): converts an object to a matrix.as.vector(): converts an object to a vector.as.factor(): converts an object to a factor.
Let’s convert my_matrix, the object we created earlier, from matrix to a data frame:
# Convert my_matrix to a data frame
df5 <- as.data.frame(my_matrix)
# Check the class of the resulting data frame (should return "data.frame")
class(df5)[1] "data.frame"☛ Exercise (15 minutes)
1. Create a data frame object calledestonia_slopecontaining the following three columns and their respective elements:
slope: Fill this column with five slope values (0, 50, 100, 200, 300)level: Assign qualitative slope categories such as flat, gentle, moderate, steep, and very steep.landuse: Provide the following landuse class for each respective slope category (agriculture, agriculture, shrubland, grassland, and forestland).
- Convert the data class of
slopevariable to afactor.
1.5. R Packages
Packages are collections of R functions, data, and compiled code in a well-defined format. The base installation of R comes with many useful packages as standard. These packages will contain many of the functions you will use on a daily basis.
More packages are added later, when they are needed for some specific purpose. These packages can be downloaded from a variety of sources, most commonly from CRAN website. The Comprehensive R Archive Network (CRAN) is R’s central software repository, supported by the R Foundation.
To install a package from CRAN you can use the install.packages() function with the name of your desired package in quotes as first argument.
# Install single package
install.packages("Package Name") # Replace ""Package Name"" with the actual package name
# Install multiple packages
install.packages(c("Package Name1","Package Name2", "Package Name3"))
# Note that Package names are case sensitive!While CRAN is still most popular repository for R packages, you will find quite a lot of packages that are only available from GitHub (from source). Furthermore, if you would like to try out the latest development versions of popular packages, you have to install them from GitHub.
To install and manage packages from remote sources like GitHub repositories or other version control systems, we need to install remote package first.
install.packages("remotes")Now you can install any package from GitHub repository by providing userName/packageName as argument to install_github() function.
# Install the package from GitHub
remotes::install_github("github_username/packageName") # replace "github_username" with the actual GitHub username and "packageName" with the name of the package you want to installBy default, install.packages() installs the most recent version of a package, but there may be cases where installing the latest version could disrupt your existing code. To install a particular package version, we can also use remotes::install_version() function.
remotes::install_version("package Name", "version")After the necessary packages have been installed, the next step is loading or importing them. This process makes the functions, datasets, and other assets offered by the package accessible for utilization within your R session. When you load or import a package, it’s typically added to the R library or environment, making its functionality accessible for your current R script or session.
R packages are stored under a directory called library in the R environment. To load a package, we use the library() function:
# Load the specified package (replace "package Name" with the actual package name)
library("package Name")The .libPaths() function is used to display the library paths where R looks for installed packages. These library paths are directories where R stores packages that can be loaded and used in your R sessions. To display the library paths where R is currently looking for packages, simply run the following command:
.libPaths()[1] "/home/geoadmin/R/x86_64-pc-linux-gnu-library/4.3"
[2] "/usr/local/lib/R/site-library"
[3] "/usr/lib/R/site-library"
[4] "/usr/lib/R/library" The above code provides two library paths where R is looking for installed packages - user-specific library path and system-level library path.
You can get the list of all the installed packages and their versions using the following command:
# List all installed packages
installed_packages <- installed.packages()
# Create a data frame with only "Package" and "Version" columns
subset_installed_packages <- installed_packages[, c("Package", "Version")]
# To view the list of loaded (or attached) packages during an R session
search()1.6. Data wrangling and manipulation
The data we get to work with will rarely be in a format necessary for analysis. The process of cleaning, structuring, and transforming raw or messy data into a more usable and structured format is called data wrangling. This often involves addressing issues like missing values, merging datasets, reshaping data, and converting data types. Data manipulation, on the other hand, is a more general term that involves filtering, sorting, aggregating, or filtering data to perform specific operations.
Data wrangling and data manipulation both refer to the process of preparing and transforming raw data and making it tidy and ready for further analysis.
1.6.1. Attribute join
Spatial data which contains geometries and non-spatial data which contains attributes are often stored separately. Combining such datasets based on keys which are columns found in both datasets is called attribute join. With a join, the variables of one dataset with the corresponding variables in another dataset are connected to each other, thus facilitating data analysis and integration.
In this lesson, we will work with two distinct datasets: the settlement dataset, which exclusively provides geometric information, and the population dataset in CSV format, which contains population-related attributes for various settlements. To effectively visualize the distribution of population across these settlements, we must join the population attributes with spatial information.
To achieve this, we will require two packages: sf and tidyverse. Tidyverse is a collection of eight core R packages designed to facilitate data manipulation, visualization, and analysis. We will be utilizing this package consistently throughout the course.
The sf package is specifically designed for handling vector data in R. It stands for “simple features” and provides a unified framework for representing and manipulating vector data. We will explore more about this package in lesson 2.
Install and load packages:
# Install packages
# install.packages("tidyverse")
# install.packages("sf")
# Load packages
library(sf)
library(tidyverse)
#Please be aware that when installing R packages, you should enclose the package names in quotation marks, whereas when loading packages, no quotation marks are needed.Let’s import the settlements data containing the geometric information and take a closer look at its contents. The st_read() function of the sf package can be used to read a vector data by specifying the path to your data.
Download the settlements data from this link and save the file in the same directory as the R script file (…/geopython2023/R_01).
# Import settlements data
geometry <- st_read("settlements.gpkg") Reading layer `geometry' from data source
`/home/geoadmin/dev/simply-general/teaching/geospatial_python_and_r/R_01/settlements.gpkg'
using driver `GPKG'
Simple feature collection with 4713 features and 1 field
Geometry type: MULTIPOLYGON
Dimension: XY
Bounding box: xmin: 369032.1 ymin: 6377141 xmax: 739152.8 ymax: 6634019
Projected CRS: Estonian Coordinate System of 1997# Determine the data class of the "geometry" object
class(geometry)[1] "sf" "data.frame"# Display the structure
glimpse(geometry) Rows: 4,713
Columns: 2
$ KOOD <chr> "9599", "1885", "3050", "2654", "9010", "1846", "1081", "4193", "…
$ geom <MULTIPOLYGON [m]> MULTIPOLYGON (((693596.1 64..., MULTIPOLYGON (((6884…The result shows that the geometry object contains two variables: geom (representing the geometric information) and KOOD (representing unique codes).
Now, let’s generate a plot using the basic R plot function:
plot(geometry$geom)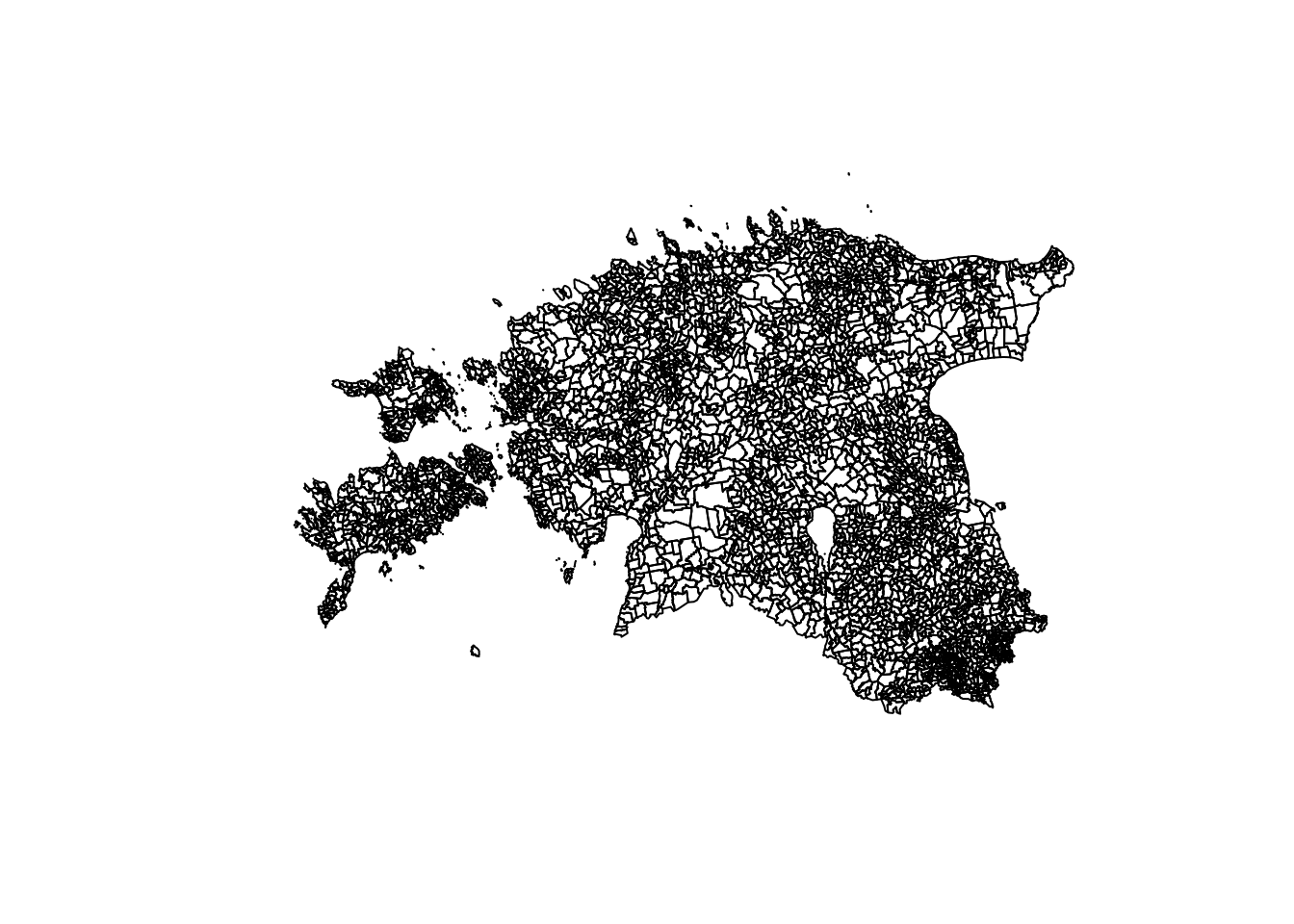
Now, let’s import the population data. To import CSV files into R, we can make use of the read.csv() function from the readr package, which is a component of the tidyverse collection.
Please download the population data from this link and save the files in the same directory as the R script file (…/geopython2023/R_01)
# Import population data
pop <- read.csv("population.csv")
#The "KOOD" variable in the population data comprises four-digit numbers.
# However, during the CSV saving process, elements starting with leading zeros were automatically removed for certain reasons.
# The next code appends leading zeros to the "KOOD" values that consist of only three digits.
pop$KOOD <- str_pad(pop$KOOD, width = 4, side = "left", pad = "0")
# Display a structured summary
glimpse(pop)Rows: 4,713
Columns: 7
$ VID <int> 66016912, 66013199, 66013544, 66013330, 66016492, 66013175,…
$ KOOD <chr> "9599", "1885", "3050", "2654", "9010", "1846", "1081", "41…
$ NIMI <chr> "Võuküla", "Holvandi küla", "Kiisa küla", "Kanassaare küla"…
$ VAARTUS <chr> "60", "46", "74", "54", "65", "501", "100", "146", "239", "…
$ STAMP_CRE <chr> "1/25/2021", "1/25/2021", "1/25/2021", "1/25/2021", "1/25/2…
$ JUHUSLIK <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ GEOKODEER8 <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…With both the spatial and non-spatial datasets available, we can now move forward and join them using a common field, denoted by KOOD. In R, there are different types of attribute joins. In our case, we will use the left_join() function from the dplyr package in tidyverse collection. The left_join returns all rows from the first data frame regardless of whether there is a match in the second data frame.
# Join pop and geometry based on "KOOD" field
pop_geom_joined <- left_join(pop, geometry, by = "KOOD")Oops, we’ve encountered an error related to the data type discrepancy in the KOOD variable. In the pop object, KOOD is stored as an integer, while in the geometry object, it’s a character. To resolve this and maintain consistency, we need to convert the data type of the “KOOD” variable in the “pop” object to character, aligning it with its data type in the “geometry” object.
# Convert KOOD from integer to character type
pop <- pop %>%
mutate(KOOD = as.character(KOOD))
# join again
pop_geom_joined <- left_join(pop, geometry, by = "KOOD")The %>% (or since R 4.1.0 the native |>) in R is called the forward pipe operator. It allows us to combine multiple operations in R into a single sequential chain of actions. It takes the left-hand side output as the first argument for the right-hand side function, streamlining data transformations and manipulations.
Now, let’s explore the joined (pop_geom_joined) dataset more thoroughly:
# Provides a structured summary of your data
glimpse(pop_geom_joined)
# colnames() retrieves the column names in your data
colnames(pop_geom_joined)
# summary() provides a summary of the main descriptive statistics for each variable in your data
summary(pop_geom_joined)
# dim() returns the dimensions (number of rows and columns) of your data
dim(pop_geom_joined)
# Checks and displays the class of your data
class(pop_geom_joined)Now, we have successfully joined the population attributes with spatial information and created a new object as a data frame. This data frame structure is ideal for our data manipulation and preparation processes. However, when it comes to spatial visualization at a later stage, it will be necessary to transform it into the sf format.
Let’s start the process of tidying up our data by deleting certain columns that are not important for our analysis. R offers different options to delete columns:
using the
$operator withNULL, commonly used for eliminating a single column. In R, the$symbol is used to access specific elements of an object, be it variables within a data frame or elements within a list.using the
select()function with a negative sign (-) preceding the variable names.
# Deleting "VID" column using the $ operator
pop_geom_joined$VID <- NULL
# Deleting multiple columns using the select() function and %>% operator
pop_geom_joined <- pop_geom_joined %>% # Overwriting the existing object
dplyr::select(-GEOKODEER8, -JUHUSLIK, -STAMP_CRE)Another useful function is rename(), which as you may have guessed changes the name of variables/columns. Renaming columns allows you to assign more meaningful and descriptive names to variables. Renaming columns in R is a straightforward process using the rename() function.
Let’s rename the Estonian column names in our pop_geom_joined object to English for common understanding:
# Create a new data frame, pop_geom_joined_renamed, by renaming columns from pop_geom_joined
pop_geom_joined_renamed <- pop_geom_joined %>%
rename(Settlements = NIMI, # rename the "NIMI" column to "Settlements,
Population= VAARTUS, # rename the "VAARTUS" to "Population
Code = KOOD) # rename the "KOOD" to "Code" Thus far, we’ve explored techniques for deleting, renaming, and selecting specific variables in the data frame. Now, we can move forward with a more in-depth analysis by focusing on the Population variable only from our cleaned dataset.
Let’s check the format of the “Population” variable within the “pop_geom_joined_renamed” object:
glimpse(pop_geom_joined_renamed)Rows: 4,713
Columns: 4
$ Code <chr> "9599", "1885", "3050", "2654", "9010", "1846", "1081", "4…
$ Settlements <chr> "Võuküla", "Holvandi küla", "Kiisa küla", "Kanassaare küla…
$ Population <chr> "60", "46", "74", "54", "65", "501", "100", "146", "239", …
$ geom <MULTIPOLYGON [m]> MULTIPOLYGON (((693596.1 64..., MULTIPOLYGON …The population variable is expected to be of a numeric data type. In our current context, it is stored as a character type, necessitating its conversion to a numeric data type. As demonstrated previously, the transformation from character to numeric is straightforward using the as.numeric() function. However, in this specific instance, we will employ the mutate function to perform the conversion and update the “Population” variable accordingly.
Mutate() is a function from dplyr which is used to create or modify columns in a data frame. Another similar function is transmute(), which is used to create or modify columns in a data frame while dropping all other columns.
# Create a new data frame 'pop_new' based on 'pop_geom_joined_renamed' and convert the 'Population' column to numeric data type.
pop_new <- pop_geom_joined_renamed %>%
mutate(Population = as.numeric(Population))
# Display a summary of the structure of the 'pop_new' data frame.
glimpse(pop_new)Rows: 4,713
Columns: 4
$ Code <chr> "9599", "1885", "3050", "2654", "9010", "1846", "1081", "4…
$ Settlements <chr> "Võuküla", "Holvandi küla", "Kiisa küla", "Kanassaare küla…
$ Population <dbl> 60, 46, 74, 54, 65, 501, 100, 146, 239, 51, 65, 55, 55, 51…
$ geom <MULTIPOLYGON [m]> MULTIPOLYGON (((693596.1 64..., MULTIPOLYGON …Let’s examine the summary statistics of the “Population” variable:
summary(pop_new$Population) Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
4.0 19.0 40.0 299.2 84.0 116153.0 282 1.6.2. Handling missing data
Handling missing data is one of the common tasks in data analysis. In R, missing values are often represented by the symbol NA. R provides various functions and techniques to identify and handle missing values.
The sum() function in conjunction with the is.na()provides the count of NAs within our object.
Let’s examine whether the Population variable contains any missing values both before and after converting its data type.
# Check if "pop_geom_joined_renamed" contains any missing values
anyNA(pop_geom_joined_renamed) [1] FALSE# What about "pop_new"?
anyNA(pop_new) [1] TRUE# Total number of NAs
sum(is.na(pop_new$Population))[1] 282The outcome indicates that there are 282 missing values (NAs) within the “Population” variable following its conversion to a numeric format. What is the source of these missing values (NAs)?
Let’s closely examine the “Population” column by using the View() function:
View(pop_geom_joined_renamed)The result indicates that there are values marked as <4, signifying population sizes that are less than 4 and have been anonymized for privacy reasons. R automatically assigns NAs to such values, resulting in the appearance of the 282 NAs.
If there’s a need to eliminate NAs from the dataset for any purpose, you can make use of na.exclude() or drop_na() functions:
# Eliminate missing values from the "pop_new" data
na_excl <- na.exclude(pop_new)
na_drop <- drop_na(pop_new)
# Check if "na_drop" contains any missing values
anyNA(na_drop) [1] FALSEUpon a thorough inspection of the NAs within the dataset, the next step involves addressing them, as their presence can potentially disrupt the analysis. In our specific case, the NAs will be addressed by substituting them with the value 2, which represents an approximate “average” for the population range that has been converted to NAs. By making this substitution, we can maintain the integrity of the overall population data while effectively.
In the next code, the coalesce() function from the dplyr package checks if the Population column has NAs and then replaces them with the 2.
# Fill missing values in "Population" column with value 2
pop_new_filled <- pop_new %>%
mutate(Population = coalesce(Population, 2))
# Check if the NAs has been successfully replaced
sum(is.na(pop_new_filled$Population))[1] 0Great! Now we have successfully handled the NAs in our data and made it ready for further analysis. Before proceeding into more in-depth analysis, let’s gain a better understanding of the Population variable in our updated object (pop_new_filled) by using basic descriptive statistics.
# Find the minimum value
min(pop_new_filled$Population) [1] 2# Find the maximum value
max(pop_new_filled$Population) [1] 116153# Calculate the standard deviation
sd(pop_new_filled$Population) [1] 3115.68# Calculate the median
median(pop_new_filled$Population)[1] 36# Generate a summary of descriptive statistics
summary(pop_new_filled$Population) Min. 1st Qu. Median Mean 3rd Qu. Max.
2.0 16.0 36.0 281.4 80.0 116153.0 # Determine the interquartile range (IQR)
IQR(pop_new_filled$Population)[1] 64# Calculate specific percentiles (25th, 50th, and 75th)
quantile(pop_new_filled$Population, probs = c(0.25, 0.50, 0.75))25% 50% 75%
16 36 80 We can also use histogram to better understand the distribution frequency of the Population data.
hist(pop_new_filled$Population)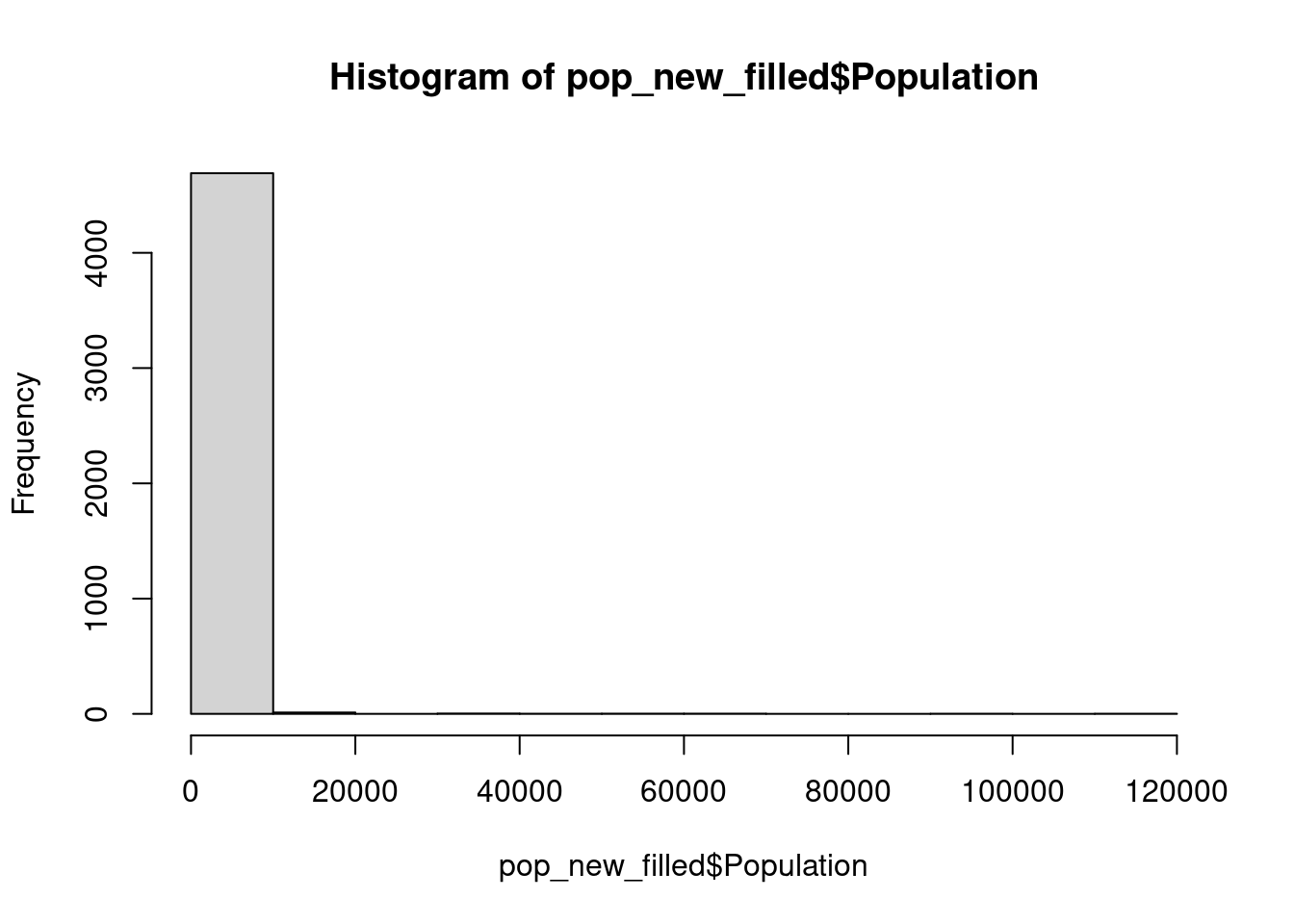
The histogram shown above depicts that a substantial proportion of the population is concentrated within the 0 to 20,000 range. Nevertheless, the extended width of the x-axis can be attributed to the presence of certain settlements in our population dataset with populations as high as 120,000.
Now, we can proceed to extract particular observations from the data frame. To accomplish this, we will employ the filter() function. Filter allows us to selectively extract rows from a data frame based on specific conditions or criteria.
The next code filters Population > 2000, and then arrange the remaining rows in descending order of Population:
# Filter and arrange population
pop_2000 <- pop_new_filled %>%
filter(Population > 2000) %>%
arrange(desc(Population))
slice(pop_2000, 1:5) # slice() function is used to select specific rows from a data frame, rows 1 to 5 in this caseWe can also use the filter() function to extract cities with a population exceeding 15000. It’s worth mentioning that in Estonia, cities are typically identified by names ending in linn.
# Filter and sort data to create a new data frame called "cities15000"
cities15000 <- pop_new_filled %>%
filter(str_detect(Settlements, "linn$")) %>%
filter(Population > 15000) %>%
arrange(Settlements)
# the "$" specifies that "linn" should occur at the end of the "Settlements" strings
# Display the "cities15000" data frame
cities15000Let’s proceed with narrowing down our data by specifically selecting three highly populated cities in Estonia (namely Tartu, Narva, and Pärnu):
# Filter the data frame to include only specific cities
top3_cities_1 <- pop_new_filled %>%
filter(Settlements == "Tartu linn" |
Settlements == "Narva linn" |
Settlements == "Pärnu linn")
# Alternatively, use "%in%" operator
top3_cities_2 <- pop_new_filled %>%
filter(Settlements %in% c("Tartu linn", "Narva linn", "Pärnu linn"))
top3_cities_1The %in% operator in R is used to check if elements from one vector are present in another vector. It returns a logical vector indicating whether each element in the first vector is found in the second vector. In our case, %in% is used to filter rows in a data frame where the Settlements column matches any of the values in the specified vector. It filters rows where the Settlements column is equal to “Tartu linn,” “Narva linn,” or “Pärnu linn.”
We can now generate a map that illustrates the population distribution across different settlements in Estonia. First we need to convert the pop_new_filled object from data frame to a spatial (sf) format using st_as_sf() function from sf package.
# Convert the data frame "pop_new_filled" to sf (spatial) format
pop_new_filled_sf <- st_as_sf(pop_new_filled, crs = 3301)
# Display a summary of the new object
glimpse(pop_new_filled_sf)Rows: 4,713
Columns: 4
$ Code <chr> "9599", "1885", "3050", "2654", "9010", "1846", "1081", "4…
$ Settlements <chr> "Võuküla", "Holvandi küla", "Kiisa küla", "Kanassaare küla…
$ Population <dbl> 60, 46, 74, 54, 65, 501, 100, 146, 239, 51, 65, 55, 55, 51…
$ geom <MULTIPOLYGON [m]> MULTIPOLYGON (((693596.1 64..., MULTIPOLYGON …Next, we can create a spatial visualization using the ggplot2 function, part of the tidyverse collection. ggplot2, short for Grammar of Graphics plot, is a widely used data visualization package offering a versatile framework to generate a diverse array of both static and interactive plots. It’s based on the Grammar of Graphics, which is a comprehensive framework for visualizing data by dividing graphs into meaningful components like scales and layers. More documentation on visualization of sf objects using ggplot2 package can be found here. Be aware that the package is ggplot2, and the function is ggplot().
# Create a ggplot object and add a "pop_new_filled_sf" layer with population data
my_plot <- ggplot() +
geom_sf(data = pop_new_filled_sf,
aes(fill = cut(Population,
breaks = c(0, 50, 100, 150, 200, 120000),
labels = c("0-50", "51-100", "101-150", "151-200", "200+"))))+
# Customize the legend and add titles
labs(fill = "Population per settlement",
title = "Population Distribution in Estonia",
subtitle = "Data source: Statistics of Estonia") +
# Apply a color scale using a palette from RColorBrewer
scale_fill_brewer(palette = "YlOrRd") + # You can change "YlOrRd" to any other palette name
# Improve the overall theme for better visualization
theme_grey() +
theme(legend.position = "right") # this is position also default position
# For more RColorBrewer options, please visit https://r-graph-gallery.com/38-rcolorbrewers-palettes.html
# Display the plot
my_plot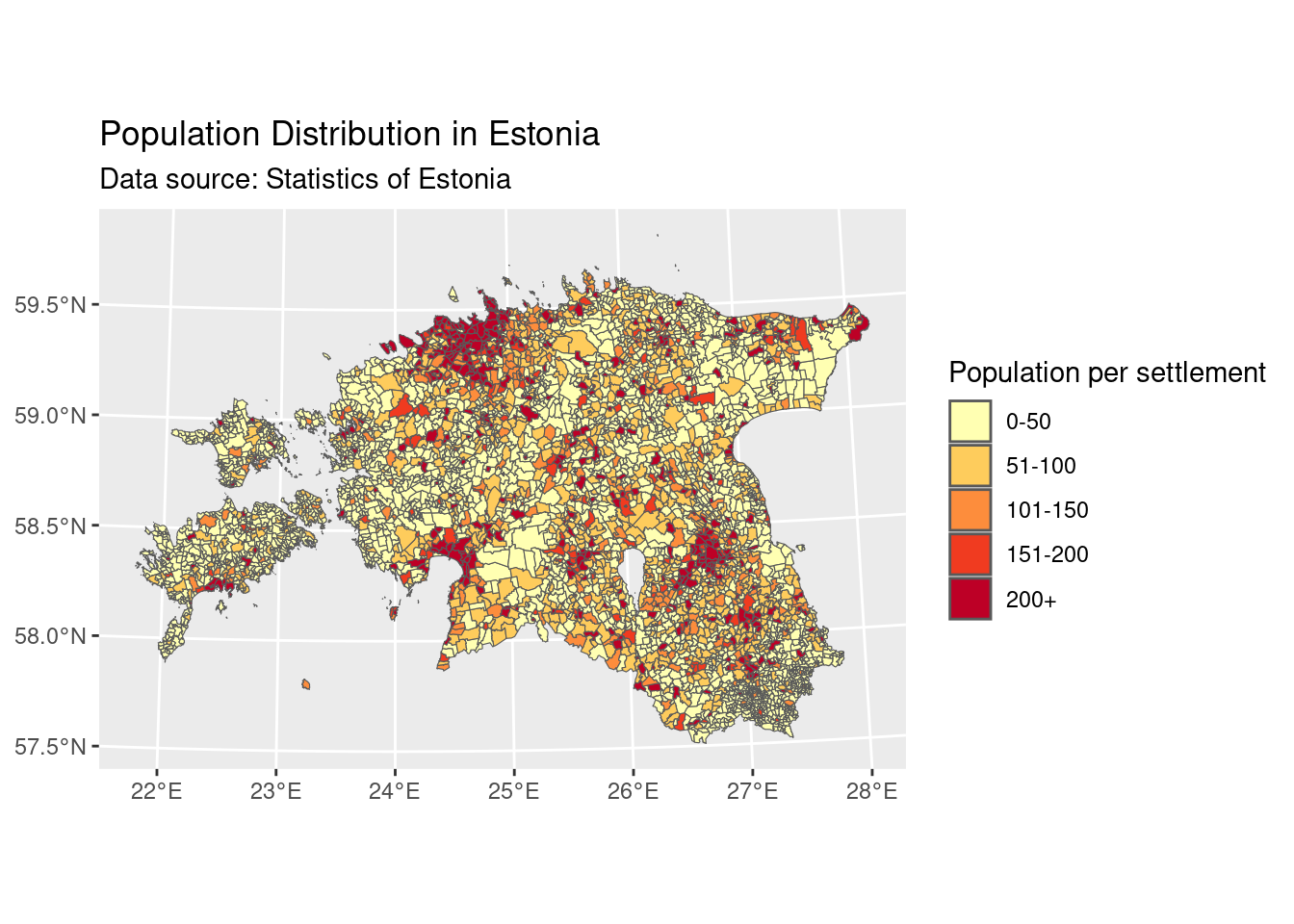
As a final step, let’s save our joined population data (pop_new_filled_sf) and the map we’ve just created (my_plot). It’s a common best practice to save the final outputs for potential future use. We can use the st_write() function from the sf package to save the sf files, and the ggsave() function from the ggplot2 package for save the ggplot maps.
# Save the sf data
st_write(pop_new_filled_sf, "population.gpkg", append = TRUE)
# Save the ggplot figure to a png file with custom width and height
ggsave("my_plot.png", plot = my_plot, width = 6, height = 4)
# Please note that this function saves the file directly to the working directory by default. If you wish to save it in a different directory, you can specify the desired path.How to get help in R?
When working with R, there are several ways to get help and find solutions to problems. Here are some common ways:
Built-in Documentation: R has extensive built-in documentation that you can access within the R environment. Thehelp()function and?help operator in R provide access to the documentation pages for R functions, data sets, and other objects, both for packages in the standard R distribution and for contributed packages. To access documentation for the standard lm (linear model) function, for example, enter the commandhelp(lm)orhelp("lm"), or?lmor??lm.R Documentation: A comprehensive collection of R packages and their documentation.
Stack Overflow: A question-and-answer website where you can search for R-related questions or ask your own. Before posting your question, make sure that it hasn’t been answered before and is well crafted, chances are you will get an answer in less than 5 min. Remember to follow their guidelines on how to ask a good question
RStudio Community: A forum for R and RStudio users to ask questions, share knowledge, and discuss R-related topics.
R Bloggers: A blog aggregator with articles, tutorials, and examples related to R programming.
R Email Lists: The R Project maintains a number of subscription-based email lists for posing and answering questions about R. Before posing a question on lists, please make sure that you read the posting guide.
- General R-help email list
- R-devel list for R code development
- R-package-devel list for developers of CRAN packages;
- R-announce email list and a variety of more specialized lists.
RPubs: A web platform provided by RStudio that allows R users to publish and share their R Markdown documents as interactive web pages
Social Media and R Forums: Join R-related groups or communities on social media platforms likeTwitter,LinkedIn, orFacebook. You can connect with other R users, ask questions, and share insights.
Reference Materials
This course does not require any mandatory textbooks. It relies on a diverse set of sources to deliver course information, and the primary references are provided below:
Geocomputation with R by Lovelace Robin, Jakub Nowosad, and Jannes Muenchow
Spatial Statistics for Data Science: Theory and Practice with R by Moraga, Paula
Geographic Data Science with R: Visualizing and Analyzing Environmental Change by Michael C. Wimberly
Spatial Data Science with applications in R by Edzer Pebesma, Roger Bivand
Spatial Analysis with R by Chia Jung, Yeh
Modern Data Science with R by Benjamin S. Baumer, Daniel T. Kaplan, and Nicholas J. Horton
R for Data Science by Hadley Wickham, Mine Cetinkaya-Rundel, and Garrett Grolemund
